
[ I am an employee of Bristol Myers Squibb. The views expressed here are my own, assuming I am real and not a humanoid. ]
In the original Blade Runner (1982), Harrison Ford’s character, Deckard, implements a fictitious Voight-Kampff test to measure bodily functions such as heart rate and pupillary dilation in response to emotionally provocative questions. The purpose: to establish “truth”, i.e., determine whether an individual is a human or a bioengineered humanoid known as a replicant.
While the Voight-Kampff test was used to establish truth for humans vs replicants, the concept of “truth” is central to neural networks used in machine learning and artificial intelligence (AI). And for AI to be effective in drug discovery and development, it is critical to ask a fundamental question: what is “truth” in drug discovery and development?
INTRODUCTION
I recently read the book Genius Makers by Cade Metz and was reminded of the long history of machine learning, neural networks, and artificial intelligence (AI). This is a field more than 60 years in the making, with slow growth for the first 50 years – AI was founded as an academic discipline in 1956 – and exponential growth in the last 10. The original mathematical framework of neural networks was created in the 50’s (perceptron), 60’s and 70’s (backpropagation), but went largely unappreciated outside of academics, as the practical applications were few and far between.…
Read full article...

[I am an employee of Celgene. The views expressed here are my own.]
In the Wizard of Oz, Dorothy clicks her heels and hopes for re-entry from her dream world by repeating, “There’s no place like home…there’s no place like home…” I often feel that many in the genetics community look at their human genetics data with the same youthful optimism as Dorothy – clicking their genetic heels and wishing “my genetic discovery will become a drug…my genetic discovery will become a drug…” But without rigor and discipline, such heel-clicking won’t overcome many of the challenges that face drug hunters along the tortuous journey from a genetic idea to a new medicine.
In this blog, I discuss a recent study on the genetics of multiple sclerosis (MS) published in Science (see here). This is a beautiful study that substantially advances the genetic landscape of patients with a devastating disease. However, the study falls short in terms of the application of human genetics to drug discovery. To chart a course for the future, I introduce the concept of mechanism, magnitude and markers (oh my!), which I refer to as the three M’s. …
Read full article...

[Disclaimer: I am an employee of Celgene. The views reported here are my own.]
I recently participated in a Harvard Medical School Executive Education course on human genetics and drug discovery (link here, slides here and here). My presentation concluded with a short discussion on emerging resources such as Phenome-Wide Association Studies (PheWAS) to predict adverse drug events and guide indication selection, and protein quantitative trait loci (pQTLs) for Mendelian randomization. In this blog, I highlight briefly our recent Nature publication on pQTLs, “Genomic atlas of the human plasma proteome” (here), which represents a new public resource for drug discovery.
Human genetic targets are endowed with favorable properties, one of which is the ability to use genetic tools for nature’s randomized control trial. Central to this concept is Mendelian randomization, a method that uses human genetic variants as an instrument to examine the causal effect of a modifiable exposure (e.g., protein biomarker) on disease in observational studies (reviewed here and recent Nature Reviews Genetics here).
Proteins provide an ideal paradigm for Mendelian randomization analysis for drug discovery, as proteins are under proximal genetic control and represent the targets of most approved drugs.…
Read full article...

A new genetics initiative was announced today: the creation of FinnGen (press release here). FinnGen’s goal is to generate sequence and GWAS data on up to 500,000 individuals with linked clinical data and consented for recall. There are many applications for such a resource, including drug discovery and development. In this blog, I want to first describe the application of PheWAS for drug discovery and development, and then introduce FinnGen as a new PheWAS resource (see FinnGen slide deck here).
[Disclaimer: I am an employee of Celgene. The views expressed here are my own.]
PheWAS
PheWAS turns GWAS on its head. While GWAS tests millions of genetic variants for association to a single trait, PheWAS does the opposite: tests hundreds (if not thousands) of traits for association with a single genetic variant. This approach is primarily relevant for those genetic variants with an unambiguous functional consequence – for example, a variant associated with disease risk or a variant that completely abrogates gene function. There are useful online resources (see here), as well as several nice recent reviews by Josh Denny and colleagues, which provide additional background on PheWAS (see here, here).
Work that originated from my academic lab represents the first example of PheWAS for drug discovery – in particular, how to use PheWAS to predict on-target adverse drug events (ADEs) and to select indications for clinical trials (see 2015 PLoS One publication here).…
Read full article...
A recent study in the New England Journal of Medicine provides genetic support for a pharmacologically validated target, BAFF, in the treatment of systemic lupus erythematosus. But can human genetics also be used to estimate the target dose and a therapeutic window?
As readers of plengegen.com know, I am constantly on the lookout for published studies that provide insight into the utility of human genetics for drug discovery and development. This past week there was a great post from Francis Collins on the role of the NIH in the discovery (in part via human genetics) and development of tofacitinib (see here), anakinra and potentially novel targets (e.g., STING) for inflammatory diseases (here). Nature Reviews Drug Discovery published a News & Analysis on PCSK9 as a “fertile testing ground for new drug modalities including long-acting RNA interference drugs, vaccines against self-antigens, CRISPR therapeutics and small molecules that control ribosomal activity” (here). New York City released information about a new public health initiative, The NYC Macroscope, which will use electronic health records (EHRs) to track conditions managed by primary care practices that are important to public health..and one day may be linked to genetic data for discovery research (that is me just speculating).…
Read full article...
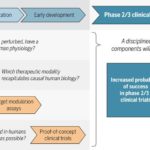
As readers of my blog know, I am a strong supporter of a disciplined R&D model that focuses on: picking targets based on causal human biology (e.g., genetics); developing molecules that therapeutically recapitulate causal human biology; deploying pharmacodynamic biomarkers that also recapitulate causal human biology; and conducting small clinical proof-of-concept studies to quickly test therapeutic hypotheses (see Figure below). As such, I am constantly on the look-out for literature or news reports to support / refute this model. Each week, I cryptically tweet these reports, and occasionally – like this week – I have the time and energy to write-up the reports in a coherent framework.
Of course, this model is not so easy to follow in the real-world as has been pointed out nicely by Derek Lowe and others (see here). A nice blog this week by Keith Robison (Warp Drive Bio) highlights why drug R&D is so hard.
Here are the studies or news reports from this week that support this model.
(1) Picking targets based on causal human biology: I am a proponent of an “allelic series” model for target identification. Here are a couple of published reports that fit with this model.…
Read full article...
I say article of the week, but I have been lazy this summer (or maybe just consumed by other things). My last “article of the week” was in May and my last Plengegen blog post was over a month ago!
By now everyone knows the PCSK9 story. Human genetics identified the target; functional work in mouse and human cells led to a mechanistic understanding of PCSK9’s role in LDL receptor recycling; therapeutic modulation was shown to lower LDL cholesterol in clinical trials; and the FDA approved drugs based on LDL lowering, with outcome trials underway to demonstrate (presumably) cardiovascular benefit. What the story highlights is that a mechanistic understanding of causal pathways in human disease is key to the success of translating targets into therapies. Further, the PCSK9 story underscores the importance of a simple biomarker (LDL cholesterol) to measure a complex causal pathway in a clinical trial.
A recent study in the New England Journal of Medicine (NEJM) provides insight into a putative causal pathway in obesity, and thus a potentially a new mechanism for therapeutic modulation. The accompanying Editorial also provides a nice perspective.
[Disclaimer: I am a Merck/MSD employee. The opinions I am expressing are my own and do not necessarily represent the position of my employer.…
Read full article...

If you could pick three innovations that would revolutionize drug discovery in the next 10-20 years, what would they be?
I found myself thinking about this question during a recent family vacation to Italy. I was visiting the Galileo Museum, marveling at the state of knowledge during the 1400-1600’s. The debate over planetary orbits seem so obvious now, but the disagreement between church and science led to Galileo’s imprisonment in 1633.
So what is it today that will seem so obvious to our children and grandchildren…and generations beyond? Let me offer a few ideas related to drug discovery, and hope that others will add their own. I am not sure if my ideas are grounded in reality, but that is part of the fun of the game. In addition, “The best way to predict the future is to invent it.”
To start, let me remind readers of this blog that I believe that the three major challenges to efficient drug discovery are picking the right targets, developing the right biomarkers to enable proof-of-concept (POC) studies, and testing therapeutic hypotheses in humans as quickly and safely as possible. Thus, the future needs to address these three challenges.
1.…
Read full article...

I attended the Mendelian randomization meeting in Bristol, UK this past week (link to the program’s oral abstracts here). The meeting was timed with the release of a number of articles in the International Journal of Epidemiology (current issue here, Volume 44, No. 2 April 2015 TOC here). This blog is a brief synopsis of the meeting – with a focus on human genetics and drug discovery. The blog includes links to several slide decks, as well as references to several published reviews and studies.
[Disclaimer: I am a Merck/MSD employee. The opinions I am expressing are my own and do not necessarily represent the position of my employer.]
Several speakers, including Lon Cardon from GSK, gave overview talks on how Mendelian randomization can be applied to pharmaceutical development. In my overview, I described important guiding principles for successful drug discovery (link to my slides here), and how Mendelian randomization (MR) is applied within this framework. In particular, I emphasized the role of establishing causality in the human system: MR is a powerful tool to pick targets by estimating safety and efficacy (i.e., genotype-phenotype dose-response curves) at the time of target identification and validation; MR is effective at picking biomarkers for target modulation; and MR provides quantitative modeling of clinical proof-of-concept (POC) studies.…
Read full article...

We held our second annual GpGx retreat last week at the Dolce Norwalk Conference Center in Norwalk, CT. The theme was “integrate and elevate”: integrate across Translational Medicine and elevate our mission to infuse cutting-edge genetics and genomics into Merck’s pipeline. What follows is a brief recount of the event.
For those who don’t like looking at photos of someone else’s family vacation, this blog post might not be for you. However, for those curious about life within pharma – read on! You might be surprised that the basic principles that create a strong community within academics or a small biotechnology company are at play within a large company like Merck. I also provide examples of Translational Medicine in action: picking the right target based on causal human biology; developing the right biomarkers based on mechanistic insight of the target; selecting therapeutic molecules (e.g., biologics) in a modality independent manner; and testing clinical proof-of-concept (POC) in small Phase Ib/IIa clinical studies.
[Disclaimer: I am a Merck/MSD employee. The opinions I am expressing are my own and do not necessarily represent the position of my employer.]
At the start of the retreat, I provided an overview on our theme: “integrate and elevate” (see slides here).…
Read full article...
This week I want to focus on the role of biomarkers in drug discovery and development, which is one of the three pillars of a successful translational medicine program (see slide deck here). The focus is on Alzheimer’s disease, based on recent articles published in JAMA. At the end of the blog you will find postings for new biomarker positions in Merck’s Translational Medicine Department.
[Disclaimer: I am a Merck/MSD employee. The opinions I am expressing are my own and do not necessarily represent the position of my employer.]
Before I start, I want to point to a few blogs that provide counterarguments to some of the optimistic opinions expressed in this blog. The first is David Dobb’s negative view on big data (here); the second on Larry Husten’s concerns about conflicts of interest between academics and industry, as it relates to a recent NEJM series (here). I will not comment further, but it is worth pointing readers to these blogs and related blogs for a balanced view on complicated topics.
I have expressed the strong opinion that what ails drug discovery and development is that we pick the wrong targets, don’t develop robust biomarkers, and we don’t test therapeutic hypotheses quickly enough in clinical trials.…
Read full article...
The primary purpose of this blog is to recruit clinical scientists into our new Translational Medicine department at Merck (job postings at the end). However, I hope that the content goes beyond a marketing trick and provides substance as to why translational medicine is crucial in drug discovery and development. Moreover, I have embedded recent examples of translational medicine in action, so read on!
[Disclaimer: I am a Merck/MSD employee. The opinions I am expressing are my own and do not necessarily represent the position of my employer.]
There is a strong need to recruit clinical scientists into an ecosystem to develop innovative therapies that make a genuine difference in patients. This ecosystem requires those willing to toil away at fundamental biological problems; those committed to converting biological observations into testable therapeutic hypotheses in humans; and those who develop therapies and gain approval from regulatory agencies throughout the world. The first step is largely done in academic settings, and the other two steps largely done in the biopharmaceutical industry…although I am sure there are many who would disagree with this gross generalization!
The term “Translational Medicine” has been broadly used to describe the second step, thereby bridging the Valley of Death between the first and third steps.…
Read full article...

Dear Mr. President,
I was very pleased to listen to your State of the Union address and learn of your interest in Precision Medicine. As I am sure you know, this has led to a number of commentaries about what this term actually means (here, here, here). I would like to provide yet another perspective, this time from someone who has practiced clinical medicine, led academic research teams and currently works in the pharmaceutical industry.
Let me start by acknowledging that I know very little about your plan, but that is because no plan has been announced. However, that inconvenient fact should not prevent me from forming a very strong opinion about what you should do. Similar behavior is observed in politics (which you know well) and sports radio (see for example “Deflate-gate”). So here it goes…
I want to clarify my definition of “precision medicine” (see here for my previous blog on how this is different from “personalized medicine”). In the simplest of terms, precision medicine refers to the ability to classify individuals into subpopulations based on a deep understanding of disease biology. Note that this is different than what clinicians normally practice, which is to classify patients based on signs and symptoms (which can be measured by clinicians as part of routine clinical appointments).…
Read full article...

At the Harvard-Partners Personalized Medicine Conference last week I participated in a panel discussion on complex traits. When asked about where personalized medicine for complex traits will be in the future, I answered that I envision two major categories for personalized therapies.
(1)Development of drugs based on genetic targets will lead to personalized medicine; and
(2)Large effect size variants will be detected in clinical trials or in post-approval studies and will lead to personalized medicine.
This answer, I said, was based in part on current categories of FDA pharmacogenetic labels and in part on how I see new drug discovery occurring in the future. But did the current FDA labels really support this view?
The answer is “yes”. In reviewing the 158 FDA labels (Excel spreadsheet here), my crude analysis found that 31% of labels fall into the “genetic target” category (most from oncology – 26% of total) and 65% fall into the “large effect” category (most from drug metabolism [42% of total], HLA or G6PD [15% of total]).
A subtle but important point is that I predict that category #2 (PGx markers for non-oncology “genetic targets”) will grow in the future. In other words, development of non-oncology drugs will riff-off the success of drugs developed based on somatic cell genetics in oncology. …
Read full article...

So, you have a target and want to start a drug discovery program, do ya? How would you do it?
When I was at Brigham and Women’s Hospital, Harvard Medical School and the Broad Institute, I presented an idea from an early GWAS of rheumatoid arthritis (RA, see here) to Ed Scolnick (former president of Merck Research Labs, now founding director of the Stanely Center at the Broad Institute, see here). In this study, we found evidence that a non-coding variant at the CD40 gene locus increased risk of RA. The first questions he asked: How does the genetic mutation alter CD40 function? Is it gain-of-function or loss-of-function? What assay would you use for a high-throughput small molecule screen to recapitulate the genetic finding?
I was caught off-guard. Sadly, I had never really thought about all of the details. At the time, I knew enough as a clinician, biologist and a geneticist to appreciate that CD40 was an attractive drug target for RA. However, I was quite naïve to the steps required to take a target into a drug screen. That simple conversation led to several years worth of work, which ultimately led to a proof-of-concept phenotypic screen published in PLoS Genetics five years later (see here).…
Read full article...

Question: What can we learn from Sputnik (see here), DARPA (see here) and disruptive innovation (see here) to invent new drugs?
Answer: The best way to prevent surprise is to create it. And if you don’t create the surprise, someone else will. (This is a cryptic answer, I know, but I hope the answer will become clearer by the end of the blog.)
My previous blogs highlighted (1) the pressing need to match an innovative R&D culture with an innovative R&D strategy rooted in basic science (see here), and (2) the importance of phenotype in target ID and validation (TIDVAL) efforts anchored in human genetics (see here). Now, I want to flesh out more of the scientific strategy around human genetics – with a focus on single genes and single drug targets.
To start, I want to frame the problem using an unexpected source of innovation: the US government.
There is an interesting article in Harvard Business Review on DARPA and “Pasteur’s Quadrant” – use-inspired, basic-science research (see here and here). This theme is critically important for drug discovery, as the biopharma industry has a profound responsibility to identify new targets with increased probability-of-success and unambiguous promotable advantage (see here). …
Read full article...

As I sought advice from colleagues about my career, I was frequently asked if I would prefer to work in academics or industry (emphasis on the word “or”). The standard discussion went something like this:
ACADEMICS – you are your own boss and you are free to chose your own scientific direction; funding is tight, but good science still gets funded by the NIH, foundations and other organizations (including industry); the team unit centers around individuals (graduate students, post-docs, etc), which favors innovative science but sometimes makes large, multi-disciplinary projects challenging; there is long-term stability, including control over where you want to work and live, assuming funding is procured and good ideas continue; your base salary will be less than in industry, but you still make a good living and there are opportunities to consult – and maybe even start your own company – to supplement income. Bottom line: if you want to do innovative science under your own control, work in academics – as that is where most fundamental discoveries are made.
INDUSTRY – there are more resources, but those resources are not necessarily under your control (depending upon your seniority); the company may change direction quickly, which changes what you are able to work on; while drug development takes 10-plus years, many goals are short-term (several years), which limits long-term investment in projects that are risky and require years to develop; the team unit centers around projects (e.g.,…
Read full article...

Genetics can guide the first phase of drug development (identifying drug targets, see here ) as well as late phase clinical trials (e.g., patient segmentation for response/non-responder status, see here ). But is there a convergence between the two areas, or pharmaco-convergence (a term I just made up!)? And are there advantages to a program anchored at both ends in human genetics?
Consider the following two hypothetical examples.
(1) Human genetics identifies loss-of-function (LOF) mutations that protect from disease. The same LOF mutation is associated with an intermediate biomarker, but is not associated with other phenotypes that might be considered adverse drug events. A drug is developed that mimics the effect of the mutation; that is, a drug is developed that inhibits the protein product of the gene. In early mechanistic studies, the drug is shown to influence the intermediate biomarker in a way that is consistent to that predicted by the LOF-protective mutations. Further, because functional studies of the LOF-protective mutations provide insight into relevant biological pathways in humans (e.g., a gene expression signature that correlates with mutation carrier status), additional information is known about genomic signatures of those who carry the LOF-protective mutations (which mimics drug exposure) compared to those who do not carry the LOF-protective mutations (which mimics those who are not exposed to drug).…
Read full article...

Are the same standards applied to genetic and non-genetic tests in clinical medicine? In a review by Munir Pirmohamed and Dyfrig Hughes (download PDF here), the authors “strongly argue that the slow progress in the implementation of pharmacogenetic (and indeed other genetic) tests can partly be explained by the fact that different criteria are applied when considering genetic testing compared with non-genetic diagnostic tests.” They provide a few compelling examples:
(1) Atomoxetine
There is no regulatory requirement to undertake clinical trials to show that the dosing recommendations for patients with, for example, renal impairment are equivalent in terms of clinical outcomes to those for patients with normal renal function. Indeed, such a stipulation would be impractical and costly, and would never be done during the drug development process, potentially disadvantaging vulnerable patient populations.
Atomoxetine, a drug widely used for attention deficit hyperactivity disorder, is metabolized in the liver by CYP2D6. The SmPC for atomoxetine states that the dose should be reduced by 50% in patients with hepatic impairment (Child-Pugh class B), as drug exposure goes up by twofold. It is also known that drug exposure is increased by a similar amount in CYP2D6 PMs; however, although the SmPC for atomoxetine mentions the effect of CYP2D6 polymorphisms, it does not mandate testing for their presence.…
Read full article...



















