[I am an employee of Celgene. All views expressed here are my own.]
What is the clinical significance of residing within the tail of a distribution for disease risk? A new study published in Nature Genetics uses a composite polygenic score to measure extremes of genetic risk(see original article here). The authors make the bold statement: “it is time to contemplate the inclusion of polygenic risk prediction in clinical care”. In this plengegen.com blog, I briefly review the paper, frame the impact of the study in terms of “long tails”, and propose how genetic tails may be used as part of a healthcare system reimagined.
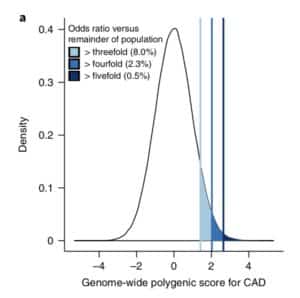
The premise of the paper is that a genome-wide polygenic score (GPS) – a composite genetic test that includes thousands and sometimes millions of genetic variants – can identify a small number of individuals from the general population that have an elevated risk. The study applies polygenic risk scores to five common diseases but spends most attention to coronary artery disease (CAD). For each disease, the increase in risk is approximately 3- to 5-fold higher among individuals at the extreme of the polygenic tail compared to those in the general population – see Figure 2a (and below) for CAD, where ~8% of the general population is at a 3-fold increase in risk based on a polygenic risk score.
Next, they compare the number of patients with an increase in risk based on polygenic risk to the number of patients with highly penetrant, rare mutations that cause monogenic disease. For familial hypercholesterolemia, approximately 0.4% of the general population have a mutation which increases risk of coronary heart disease by approximately 3-fold. They find that the population prevalence of individuals at 3-fold risk based on polygenic score (~8% of the population) is 20-fold higher than the population prevalence based on rare mutations (~0.4% of the population). [See Footnote #1 on social media criticisms of the study.]
The abstract concludes: “We propose that it is time to contemplate the inclusion of polygenic risk prediction in clinical care.”
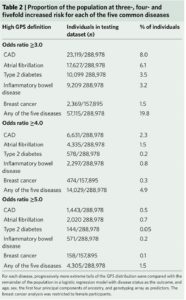
What I find fascinating about the study is that it emphasizes disease risk in the tails of a population distribution. In doing so, the study reminds us that we are all tails of some health-disease distribution, even if we don’t know what “our disease” risk profile is. In the Khera et al study, approximately 20% and 1.5% of participants were at ≥3-fold and ≥5-fold increased risk for at least 1 of the 5 diseases studied, respectively (see Table 2).
But are genetic tails actionable? We generally accept that single gene mutations responsible for Mendelian diseases are clinically actionable, as these mutations are often highly penetrant, provide a definitive diagnosis, and occasionally lead to a therapeutic intervention. As in the Khera et al paper, however, not all monogenic disease mutations are highly penetrant. In addition, as putative pathogenic mutations are uncovered via population-based sequencing, the true penetrance of these mutations will likely drop. For these less penetrant monogenic mutations (e.g., mutations that cause coronary artery disease in patients with familial hypercholesterolemia), if the same genetic risk is contributed by a more complex polygenic score but the risk prediction result is the same, should it matter if it is a simple, single gene score or a more complex polygenic score?
Although this is one way to frame the question, I think another approach is more forward-looking. It is not to consider genetic tails in the world we inhabit today but in the world in which we will live in the very near future. Consider the following argument:
- 1. Tails matter. Long tails drive innovation (see Footnote #2 below). We all distribute along a tail of disease risk, even if we don’t yet know that disease is exactly. Intuitively, we try to figure out our disease risk profile by asking basic questions. Which diseases run in my family? What ailments have affected me in my lifetime? What does my doctor tell me about my health history and future disease risk? Which diseases are prevalent among my socioeconomic peers? Which diseases am I hearing about on the news? We try and integrate this information to derive a personalized composite risk of disease.
The problem is, the human brain is terrible at integrating complex information and predicting risk! We often over- or underestimate risk, depending upon our environment. [If you want to read more about this topic, I suggest Nate Silver’s The Signal and the Noise : Why Most Predictions Fail – but Some Don’t (here) or Michael Lewis’ The Undoing Project: A Friendship That Changed Our Minds (here).] Therefore, we need a more quantitative approach to predicting our disease risk profile.
Moreover, I don’t think anybody is satisfied with the current state of conventional risk prediction. There are some useful tools, to be sure, but there has to be a better way to predict disease risk.
Thus, we need better tools to predict who is at a marked increased risk of developing disease, i.e., who is in the tail of the distribution. I don’t think there should be much argument against this conclusion.
2. Some tails are actionable. But even if we are part of a tail of disease risk, and even if genetics contributes to that risk prediction algorithm, does it matter to our long-term health? After all, some tails are actionable while others aren’t. Compare and contrast two disease: coronary artery disease (CAD) and rheumatoid arthritis (RA).
High cholesterol is a major risk factor for coronary artery disease. Decades of research have established a causal relationship between LDL cholesterol and risk of CAD. Pharmaceutical companies have developed several therapies that lower LDL cholesterol by distinct mechanisms. Each of these therapeutic approaches reduce risk of CAD. By extension, it is not unreasonable to posit that someone at genetic risk for CAD who has an elevated cholesterol level – even if elevated cholesterol does not meet current guidelines for treatment – would benefit from cholesterol lowering therapy. Thus, a genetic risk score for CAD is actionable because there are pharmacological interventions that are beneficial. [There are also lifestyle interventions that are beneficial (e.g., exercise, diet, smoking), but we all should be doing these lifestyle interventions already.]
In contrast, rheumatoid arthritis (RA) represents an example where identifying those at increased risk is not clinically actionable, at least today. Back in 2010 we found a simple genetic risk score added to a clinical model greatly improve AUC for predicting risk of RA from 0.63 to 0.75, resulting in 3-fold increase for 10% of the population (see Figure 1 & Figure 2 in this Annals of Rheumatic Disease publication). We extended our method to polygenic scores, with modest increase in predicting risk when thousands of variants were included in a model (see here for Stahl et al Nature Genetics 2012). I was a practicing rheumatologist at the time of these studies, and I would have conversations with my clinical colleagues about what we would do with this information, should it be available to us. Unlike LDL-lowering therapy and other lifestyle interventions to decrease risk of CAD, there was no obvious intervention available to decrease risk of RA. The only lifestyle intervention was to counsel against smoking, as smoking is a risk factor for RA. But, duh, every physician already counsels against smoking, which should be done regardless of a genetic risk score. Thus, a genetic risk score for RA risk was not clinically actionable. [A personal footnote: this observation was one reason I decided to move from academics into the pharmaceutical industry – see here.]
Thus, at least today, some tails are clinically actionable (LDL-lowering therapies to protect against CAD) while others are not (genetic risk of RA). But this considers the state of health care today. What about the future?
3. Tails will create new markets. I am a firm believer that as human genetics is incorporated into clinical care, we will approach the problem of health care differently. That is, technological advances such as polygenic risk scores, mobile devices, deep molecular phenotyping, artificial intelligence, etc. will create a new way to think about delivering health care. And with a healthcare system reimagined will come new ways to make information “actionable”. Or put another way, a newly imagined health care system will lead to new opportunities, much in the way that technology to enable autonomous driving has created new markets for the auto industry (see recent WSJ article here).
Here are a few bullet points to support the view that our health system is on the trajectory towards this re-imagined state (see also Footnote #3):
• Geisinger Healthcare is now incorporating genetics into every day clinical care (see here).
• Clinical trials in RA and other autoimmune diseases are evolving to include early interventions to prevent disease (see here).
• Regulatory authorities are working to modernize clinical trial design (see Scott Gottlieb’s July #SundayTweetorial here and WSJ article here).
• Technological advances continue to push the concept of “humans as model organisms” (see Science perspective here).
• Technology companies such as Amazon continue to invest in the healthcare market (see here).
• Investment dollars are flowing into health care delivery systems such as the primary care group One Medical (see here) or Mt. Sinai’s Lab100 (here).
One practical application of this future “clinically actionable” state is targeted recruitment for preventative clinical trials. Today, it is very challenging to identify those in the tails of distribution for disease risk who may benefit in the near term from a preventative intervention. As healthcare systems evolve to incorporate polygenic scores, at risk individuals will be identified as part of routine care. These individuals can be rapidly recruited into a prevention clinical trial.
For RA and other autoimmune diseases (e.g., celiac disease, type 1 diabetes, inflammatory bowel disease), for example, a future clinical trial may include patients identified via a polygenic risk score, family history, environmental risk, and other clinical features collected as part of routine care. Subjects could be randomized to a treatment vs standard-of-care cohort. Molecular (e.g., autoantibody seroconversion, inflammatory markers) and clinical endpoints could be followed for evidence of disease initiation and/or progression. With the right regulatory framework, these real-world endpoints may be sufficient for regulatory approval (see Pharmacology & Therapeutics review here).
How long will it take for this healthcare transformation? While there are many moving parts, components are being implemented today, as exemplified by the examples above. I believe the next 5-10 years will see substantial progress, given the confluence of new technologies such as polygenic scores and the burning platform to reimagine healthcare. Shortly thereafter we will reach a tipping point, much in the way the APARNET of 1960’s led to the internet of the 1990’s. Within 20-30 years we will look back on our healthcare system and reminisce in the way we do today about computers and digital technology.
[Editorial note: I was recently reminded that the pace of adoption varies greatly across society. I tried to teach my 82-year old father to text using his old LG flip phone (see photo below). He complained about the challenges of texting while refusing to upgrade to a more modern device!]
In conclusion, I believe polygenic risk predictors will become one of many technologies that will drive the re-imagination of our health care system. The study by Khera et al provides a compelling study of five common diseases, with actionable interventions in those at risk for coronary artery disease. These polygenic scores, especially when combined with conventional risk factors, will help identify those in the tails of the health-disease distribution. Together, our future healthcare system will create new approaches to make information actionable, including targeted recruitment for clinical trials, thereby improving lives of patients.

Appendix
Footnote #1: Social media criticisms of the paper. The first criticism is that that the polygenic risk scores have not been replicated. This is not true. The Khera et al study provides independent replication via the UK Biobank phase 2 – see Table 1 below, far right columns. Moreover, similar findings have been identified by others. Thus, the GPS defined by Khera et al represents an independently validated risk score.
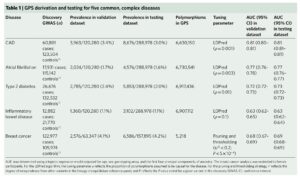
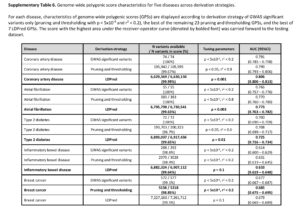
The second criticism is that adding additional variants to the risk score beyond those that are established, genome-wide significance variants does not add much to risk prediction. While this may be a valid criticism, the point is that GPS do add 1- to 2-points on the AUC for each of the 5 diseases tested. See Supplementary Table 6, below, as an example; compare AUC from top line vs AUC in bold for each disease. Given the option, I would prefer to have a model that has higher predictive value. Wouldn’t you? Sure, the increase is modest, but it is an increase in predictive value nonetheless. For more on this debate, see @cecilejanssens tweetorial here.
A third criticism is that the monogenic diseases used as benchmarking in this paper – familial hypercholesterolemia and a mutation in HNF1A that increases risk of type 2 diabetes – are not the most appropriate benchmarks. Most mutations that cause monogenic diseases have a much higher increase risk of disease. The counterpoint to this criticism is that many “pathogenic” variants responsible for monogenic diseases likely overestimate risk, as these variants were ascertained in families with disease. As unbiased genome-wide sequence is performed, it is very likely that the true penetrance of these putative pathogenic mutations will decrease.
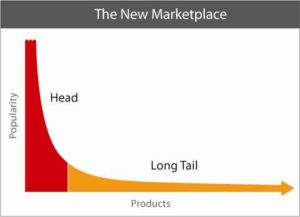
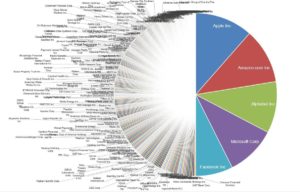
Footnote #2: Long tails drive innovation. The concept that long tails drive innovation is not new. Chris Anderson’s 2006 book, The Long Tail, describes how niche products create unique markets. Nassim Taleb’s 2007 book, The Black Swan, focuses on the extreme impact of rare and unpredictable outlier events (see @DShaywitz‘s WSJ book review here). More recently, a Collaborative Fund blog provides a compelling argument that many of the most important events in our lives are the result of long tails. “Long tails drive everything. They dominate business, investing, sports, politics, products, careers, everything. Rule of thumb: Anything that is huge, profitable, famous, or influential is the result of a tail event.” As an example, the top 5 companies from the S&P 500 make up half of the value, i.e., the top 1% of companies make up 50% of value (Figure here and below). Thus, tails are an important part of advancement in society – ignore tails at your own risk!
Footnote #3: Biotechnology investment. While not directly related to long tails or polygenic risk scores, I point readers to this blog on value creation within early-stage biotech companies. Many of the examples are technology companies, including those that were early adopters of genetic and genomics. Another resource is this Bio Report podcast on investment dollars flowing into life science companies at the nexus of information technology, biotechnology and healthcare.

