When I last wrote about AI on this blog three years ago, I spoke of it being a tool with the potential to transform scientific discovery, but the application I described was primarily theoretical. For AI to be a meaningful tool in R&D, I argued, we needed better sources of “truth” – better data sets that AI tools could query and learn from over time – and technology capable of integrating multiple steps into a semi-automated system. My message was that AI-enabled drug discovery was coming…someday.
Fast forward to 2025, and that someday is now.
We’ve seen an explosion in the availability and capability of AI tools. Just 10 months after I wrote about the theoretical possibilities of AI in biopharma, OpenAI debuted ChatGPT. Shortly after that, we saw the rollout of Microsoft Copilot and Meta AI. We now have immense computational power at our fingertips, with programs specifically designed to query biological problems. Combined with the ingenuity of skilled scientists, who can define the research problem and generate curated datasets that will enable solutions, AI has become an important and practical tool that is helping researchers accelerate discovery (link to Google DeepMind podcast on this topic here; link to a start-up’s pragmatic journey of AI in drug discovery here).
In this blog, I want to talk about how scientists at Bristol Myers Squibb (BMS) are harnessing AI tools to solve complex biological problems and support our R&D principles to achieve sustained, top-tier R&D productivity. AI isn’t changing what we do, which is discover, develop, and deliver transformational medicines to patients, but it is accelerating how we do it. (For those who prefer podcasts, I uploaded this written blog into an AI-podcast generator, which you can listen to here.)
Our framework
If you are a regular reader of this blog, you are familiar with the three research principles I believe are key to delivering transformational medicines and increasing R&D productivity: establishing causal human biology (CHB), matching modality to mechanism (MMM), and having a clear path to clinical proof of concept (P2PoC). If you need a refresher, you can check out my “Bullseye, Aim, Fire!” blog, which goes into a bit more detail. (Stay tuned for a blog on P2PoC…which is a new favorite topic of mine!)
We have evolved that framework further to include two additional principles: accelerating full development and maximizing market access. Taken together, these five end-to-end R&D principles will allow us to pursue novel targets with transformational potential and generate programs that have an increased probability of success in clinical development.

Our R&D teams at BMS are utilizing AI tools to support each of these pillars. For the purposes of this blog, however, I’m going to focus on the first three pillars and explain how we are using AI to accelerate and enhance our work in Research.

Causal human biology: from the reductionist to the physiologically relevant
Understanding causal human biology is essential in our quest to achieve our R&D ambitions. We must understand the cause-and-effect relationship between a biological target and disease, and how manipulating that target will impact human physiology.
Much of what has occurred in this space to date is based on human genetics. We now have vast amounts of data on human biology due to improvements and availability in sequencing technologies, which have unleashed an “omics” boom. However, making causal inferences from these human datasets is increasingly complex.
This is where scientists can put AI to good use. Through a mix of in vivo, in vitro, and in silico approaches, our scientists are using AI to inform our understanding of causal human biology.
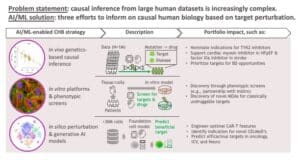
Using in vivo genetic and clinical data, our scientists are using predictive models to guide target identification and indication selection for a wide variety of diseases. We now have AI/ML capabilities that have been trained on millions of genetic samples and can predict the probability of success for various targets and novel therapeutic hypotheses. We are also using in silico modeling using generative AI to search through a massive number of potential perturbations efficiently and identify those most likely to yield a beneficial effect in specific clinical settings.
One of the most exciting ways we are using AI/ML is through AI-enabled phenotypic screens, which have become an important complement to genetics-based causal inference. Using in vitro human cell and organoid models, we are using AI to identify targets and define mechanistic details, providing a richer readout of cellular features.
One example of this approach is our collaboration with insitro, where we are leveraging AI/ML-powered phenotypic screens using induced pluripotent stem cell (iPSC)-derived disease models for neurodegenerative disorders. Neurodegenerative diseases often have heterogeneous genetic causes, but the shared pathological and clinical features suggest there are likely shared cellular and molecular events across patient populations. Using AI/ML-enabled computer imaging, we are working to link cellular models to specific familial and sporadic patients and identify disease phenotypes and mechanisms. With insitro, we are also using a novel, AI/ML-enabled technology called Pooled Optical Screening in Human Cells (POSH) to identify genes that modify neurodegenerative diseases. Through this collaboration, we are discovering novel targets that could be used to develop treatments for diseases with high unmet need. To learn more, see here for a LinkedIn post that contains a cool video on this application in the context of our partnership in amyotrophic lateral sclerosis (ALS).
How does this work, exactly? AI/ML tools can perceive subtle differences in cells that the human brain cannot. Think of it like a “spot the difference” cartoon, except instead of identifying the differences in four or six cartoons, which humans can do relatively easily, AI is perceiving differences across thousands of samples. But it’s beyond just pattern recognition. Combined with our in vivo analyses, AI and ML will allow us to draw mechanistic insights from a vast amount of human genetic and clinical data. As insitro CEO Daphne Koller recently explained in an interview with the AP: “You can measure systems like proteins and cells with increasingly better measurements and technology. But if you give that data to a person, their eyes will just glaze over because there’s only so many cells someone can look at and only so many subtleties they can see in these images.”

However, the most important factor in all of this is the involvement of our scientists. These technologies are people-driven and patient-focused, and they are only as good as the people using them and the data they feed into these predictive tools. For AI to be useful, we need to continue investing in data generation by skilled scientists, whose broad perspective and understanding of unmet patient needs outstrips AI. Machines don’t generate innovation – people using machines do. Or as described in a Harvard Business Review article, “AI Won’t Replace Humans — But Humans With AI Will Replace Humans Without AI” (here).
Matching modality to mechanism: sorting through a smaller haystack
The second pillar of our R&D framework – the “aim” in “Bullseye, Aim, Fire” – is matching modality to mechanism. Once we have identified the biological mechanisms driving a disease, we need to identify the correct molecules and modalities that will perturb this mechanism in a way that is beneficial to patients. (For a recent example of matching modality to mechanism, see this press release describing BMS’ partnership with BioArctic on a brain shuttle-enabled anti-amyloid therapy for Alzheimer’s disease.) However, molecular design is often slowed by the need to translate large, complex datasets into actionable hypotheses.
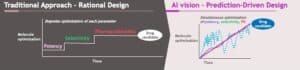
This is where AI can provide an immense benefit to researchers. Historically, researchers would use a funnel approach to identify leads, starting with a large pool of lead candidates and using a “screen and see” approach to look for binders and molecules with strong bio-performance. From there, scientists would move further and further into the funnel, as potential candidates were eliminated, and others were advanced.
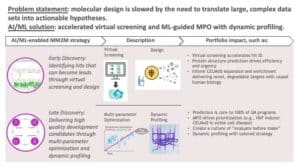
AI/ML allows researchers to short-circuit that funnel. At BMS, we call this our “Predict First” approach, which combines human intuition and computation to deliver high-quality development candidates. We use this Predict First approach across our entire small molecule portfolio, and increasingly across our large molecule portfolio, underscoring our emphasis on moving forward candidates that we predict will be successful and impactful for patients.
As part of Predict First, our colleagues in early discovery are using AI/ML in virtual screens to more quicky identify chemical and biological compounds that can become lead candidates. As our Discovery and Development Sciences leader Michael Ellis says: “Virtual screens won’t find a needle in haystack – at least not yet – but they will reduce the size of the haystack so that the needle is easier to find.”
In late discovery, researchers are using multiparameter optimization (MPO) with dynamic profiling to accelerate the identification and design of potential leads (see this bioRxiv preprint from BMS scientists). Without AI, optimizing leads is often like a game of whack-a-mole: you optimize bioavailability, but then up pops a safety issue, and then when you address the safety issue, up pops another problem. AI-enabled MPO allows you to optimize all parameters simultaneously, saving researchers valuable time. It’s like designing all aspects of Formula 1 racecars at once – optimizing tires, engine power, torque, and chassis materials in one go – and then racing your five most promising models against each other to see which is the winning design.
A great example of this in practice is our protein degrader being developed for use in sickle cell disease (SCD), a condition for which there is significant unmet need. Prior research has shown that elevating fetal hemoglobin (HbF) can ameliorate SCD-related symptoms, but many of the targets that would allow for HbF elevation were historically considered undruggable.
However, in data shared last year at the American Society of Hematology (ASH) Annual Meeting, our teams showed how we used AI to pursue these undruggable targets. Our researchers performed a phenotypic screen that found targeting two of HbF’s transcriptional repressors – zinc finger and BTB domain containing 7A (ZBTB7A) and widely interspaced zinc finger protein (WIZ) – resulted in the most effective induction of γ-globin and the highest levels of HbF. AI-driven MPO was then used to identify a compound that would optimize the degradation of ZBTB7A and WIZ for clinically meaningful HbF elevation. As BMS scientist Neil Bence described in a recent interview: “In preclinical models, it (the therapeutic protein degrader) induces fetal hemoglobin to levels that are predicted to offer functional cure potential.” The asset is currently under clinical investigation for patients with SCD.
AI is taking us to the ledge of the undruggable through the powerful intersection of science and technology, allowing us to develop treatments for patients we were previously unable to help.
Path to clinical proof of concept: ensuring more shots on goal
Ultimately, none of the work described above will matter to patients if we don’t have a clear path for these potential medicines to go from the bench to the bedside. AI can help us do this in a few ways.
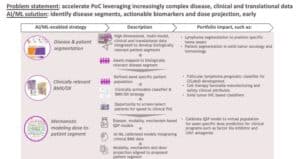
One application is patient segmentation. We know from experience that not all patients respond to standard-of-care treatments the same, but we don’t always understand why. Scientists can use AI to analyze large datasets to establish molecular profiles, uncover biological disease insights, and potentially map assets to biologically relevant disease segments.
Our researchers recently demonstrated this in a paper in Nature Communications in collaboration with scientists at the Mayo Clinic. Anita Gandhi and other BMS scientists partnered with the Mayo Clinic to build a large dataset of more than 1,000 patients with diffuse large B-cell lymphoma (DLBCL). Working with members from our Informatics and Predictive Sciences team, scientists performed an unsupervised analysis using AI to determine how DLBCL patients cluster based on their molecular profiles. They identified seven patient clusters, one of which – a group they called A7 – was found to have poor prognosis. Further analysis showed that tumors in this high-risk cluster exhibit decreased MHC expression and a less immune infiltrated tumor microenvironment. Using pre-clinical models, the researchers provided a mechanistic rationale for overcoming the low immune infiltration phenotype of the A7 cluster by using Ikaros/Aiolos protein degraders – an approach that would also work for patients in the six other clusters as well.
The concept of patient segmentation as described above isn’t new, but our scientists now have more robust datasets and more computational power, which will allow for both the precise classification of patient clusters and the identification of biomarkers. If we can identify molecular profiles of high-risk patients, we can not only ensure we are developing treatments that are effective in broad patient populations inclusive of high-risk groups, but we can also better identify high-risk patients in the clinic and – maybe someday – develop precise treatments for these subgroups. This will allow us to get the right treatment to the right patient at the right time.
Additionally, researchers can integrate AI with mechanistic computational modeling to predict how a drug will react in the human body — like a sneak peek into the future of a clinical trial. These tools can help us predict dose responses, tissue distribution and safety profiles more accurately. This integrated strategy not only accelerates our path to PoC but also enhances our ability to make data-driven decisions that improve our probability of success in the clinic. It gives us more shots on goal, so that we can increase the probability a drug will reach patients and provide them with transformational outcomes.
A future of collaborative hybrid intelligence
I opened my last AI-focused blog with a reference to Blade Runner, the 1982 Sci-Fi movie starring Harrison Ford. Like many Hollywood films about AI, Blade Runner portrays a dystopian future in which technological advancement leads to human estrangement, violence, and an existential battle for the soul of humanity. It’s all very…dark.
When it comes to AI in drug discovery, that dystopian view is all wrong. To be clear, I do not think AI will usher in a drug discovery utopia either. AI and ML are practical tools that scientists can use to augment and accelerate the discovery process. In the same way that the development of email changed the way we communicate with colleagues, AI will change the way we work. But companies did not evolve to become “email companies”; they became companies made up of people who use email. Similarly, I think we will see an evolution where biopharma companies will be made up of people and teams who use AI.
At BMS, our vision for AI/ML in research is one of “Collaborative Hybrid Intelligence.” The people are key. Scientists define the problem statements and propose the AI solutions. They identify the datasets needed to solve complex biological challenges and collect the human data to fulfill that need. The data serve as the foundation for generating insights using AI tools, but the talent surrounding the data decide how to use those insights and make the key decisions that drive innovation.
We are now in an era where we have shrunk the space between computational and molecular biologists, and we are only at the beginning. We are bringing AI and ML to life at BMS through the right mix of talent, culture, and data, and unlike the dark environs in Blade Runner, I believe the future of AI in biopharma is bright.


